Genomic instability
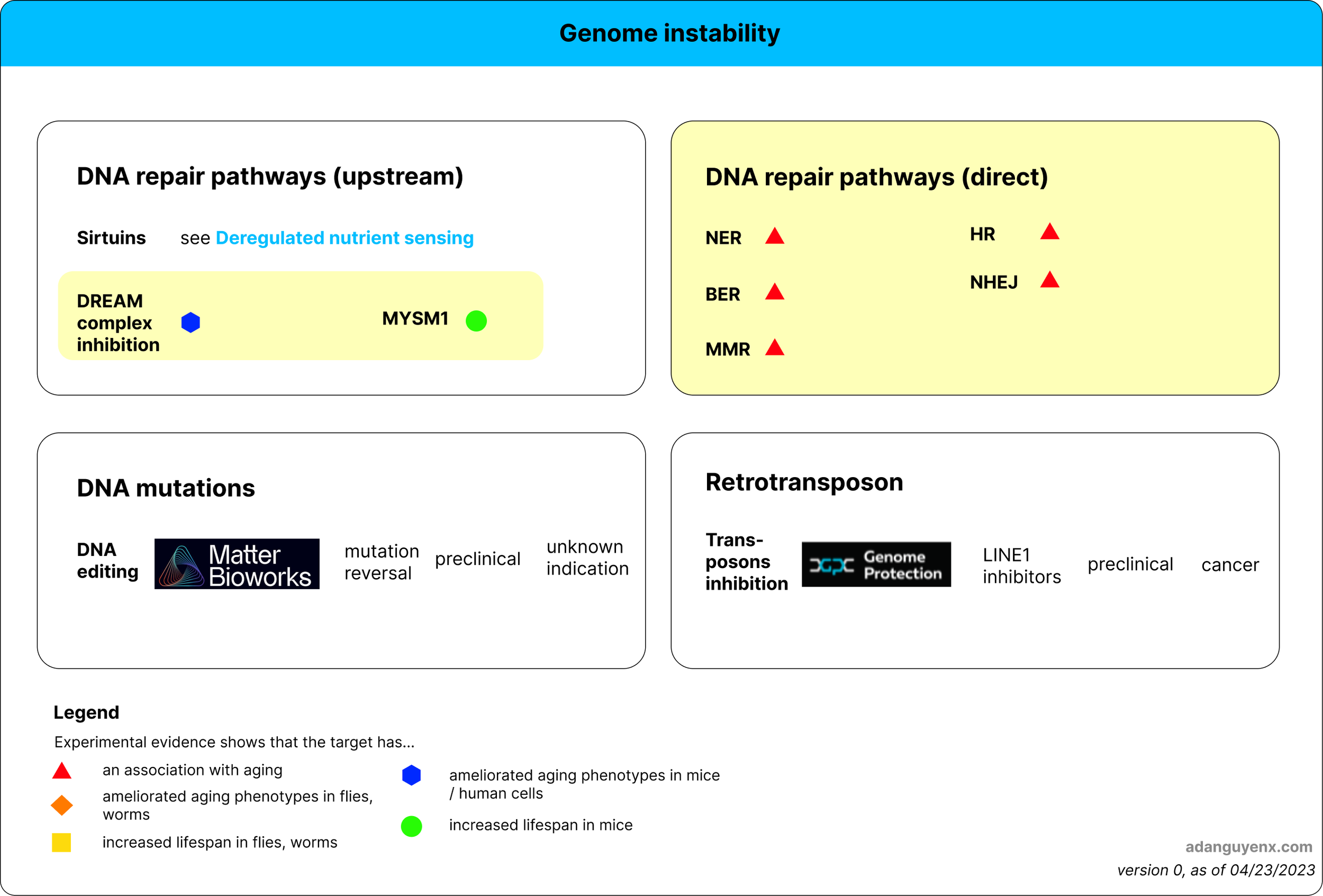
Figure 1: Therapeutic approaches for genome instability. Areas with yellow highlight indicate target(s) not explored by existing companies. Company logo is followed by description of their approach, stage of development, and disease(s) they target.
Genome instability occurs as a result of defective DNA damage repair, somatic mutations, and retrotransposons. To address genome instability, targeted approaches can be employed to address each of these contributing factors.
DNA damage repair pathways
The genome is susceptible to damage from both endogenous and exogenous factors, so DNA damage repair is an evolutionarily conserved process that has developed very early on. The cell's machinery employs various methods to repair DNA damage, including nucleotide excision repair, base excision repair, and homologous recombination (Figure 2). Each of these pathways involves a series of intricate steps and numerous proteins.
Figure 2. DNA repair pathways (source: Vijg 2021)
With age, DNA repair capacity gets worse, leading to genome instability. To address this issue, one can enhance DNA repair through targeting upstream factors or directly upregulating proteins in the repair pathways.
1.1. Target upstream factors that can enhance DNA repair
Sirtuins and NAD+
NAD+ serves as a substrate for various proteins, such as PARPs, a family of DNA repair enzymes, and sirtuins, which are involved in regulating different DNA repair pathways (Lagunas-Rangel 2019). As a result, NAD+ availability and sirtuin activation may influence DNA repair capacity.
Although NAD+ repletion has been demonstrated to extend the lifespan of mice (Zhang et al. 2016), its involvement in other aging-related processes like mitochondrial dysfunction and metabolism means that its effects may not be exclusive to DNA damage repair. Similarly, sirtuins have been controversially linked to lifespan extension, but they also participate in other pathways.
Several companies, as described in the "Deregulated Nutrient Sensing" section, are currently developing NAD+ supplements and sirtuin activators. Since these proteins are involved in other pathways, these companies may not necessarily be considered primary DNA damage repair companies.
DREAM complex inhibition
Another upstream regulator of DNA damage repair is the DREAM complex. A recent paper from Bujarrabal-Dueso et al. (2023) shows that this complex, which is regulated by DYRK1A protein kinase, is responsible for repressing DNA repair genes. The use of DYRK1A inhibitors to inhibit the DREAM complex provided DNA damage resistance in human cells and reduced DNA damage and apoptosis in the retinas of progeroid mice with Ercc1 deficiency. Pharmacological inhibition of DREAM complex therefore presents a promising and commercially unexplored approach to address genome instability.
MYSM1
MYSM1 is a deubiquitinase enzyme that specifically targets histone 2A (H2A) and is involved in the regulation of a range of cellular functions. Tian et al. (2020) found that MYSM1 enhances DNA repair through the homologous recombination (HR) pathway. Additionally, the study showed that when 16-month-old mice were treated with adeno-associated virus serotype 9 (AAV9) carrying the MYSM1 gene (AAV9-MYSM1) for 6 months, the mice experienced an extension of lifespan and multiple health benefits, including the prevention of hair loss, a reduction in vacuolar alterations, and decreased inflammation in key organs such as the kidney, heart, and liver. MYSM1 is another potential target for genome instability that no company is currently exploring.
1.2. Directly upregulate DNA damage repair pathways
Directly upregulating DNA damage repair is harder than targeting upstream factors. The process involves multiple proteins working in coordination, so overexpressing a single protein can actually harm rather than improve repair (Shaposhnikov et al. 2015). For instance, hyperactivation of PARP1, a key enzyme in the repair of single-strand DNA breaks, has been associated with aging, abnormal metabolism, and neurodegeneration (Maynard et al. 2015).
To date, the link between longevity and DNA damage repair pathways is primarily correlative, and efforts to directly enhance DNA repair pathways have not produced beneficial outcomes. Consequently, no companies are currently pursuing this area.
2. Somatic mutations
An alternative strategy to address genome instability is to focus on the downstream effects of DNA damage, specifically somatic mutations. Somatic mutations can arise from DNA damage that is not adequately repaired and have been implicated in the aging process, as demonstrated by Cagan et al. (2022).
Reversing somatic mutations could be a promising strategy, though its impact on lifespan extension is yet to be determined. Matter Bio is a company pursuing this approach.
3. Retrotransposons
Besides DNA damages, another factor that affects genome instability is retrotransposons. Retrotransposons are mobile genetic elements that can replicate and insert themselves into different locations within the genome. Cells have evolved mechanisms to suppress retrotransposon activity and maintain genome stability. However, these mechanisms may become less effective with age, leading to increased retrotransposon mobilization and associated effects on aging.
LINE1 in particular is the most abundant retrotransposon that makes up 17% of human DNA (Beck et al. 2010). Depletion of LINE1 RNA using antisense oligonucleotides has been shown to reduce the expression of senescence-associated genes in human cells and increased lifespan in a mouse model of Hutchinson-Gilford progeria syndrome (Della Valle 2022), showing that it could be a promising therapeutic strategy for aging. Genome Protection is a longevity biotech company currently working on the development of LINE1 inhibitors, initially exploring their use in the treatment of cancer.
Overall, genome instability is one of the least popular areas of focus in the “Reset and Repair” approach, with Matter Bio and Genome Protection being the only canonical companies in this area. One reason for this is the lack of available tools for directly enhancing DNA repair. As a result, there is currently no established link between improved DNA repair and extension of lifespan. Moreover, determining clinical indications in this field can be difficult, as genome instability may not be significant enough to target as the sole factor in indications beyond progeria or cancer.